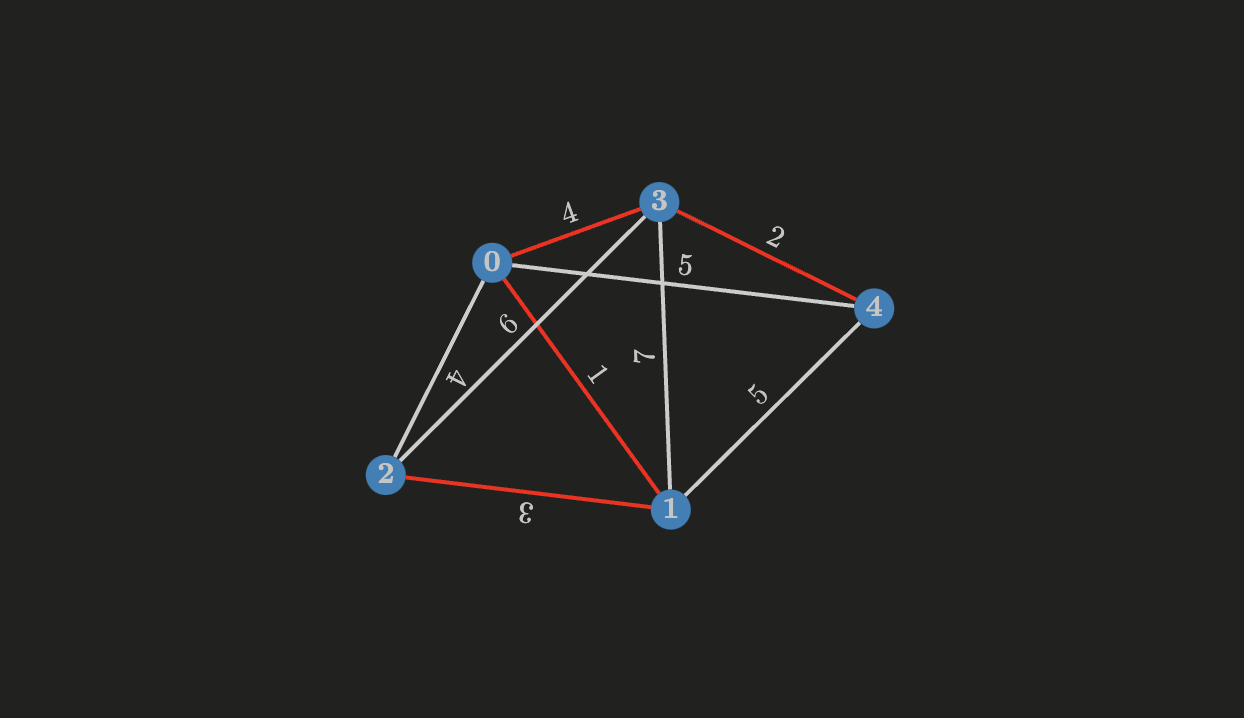
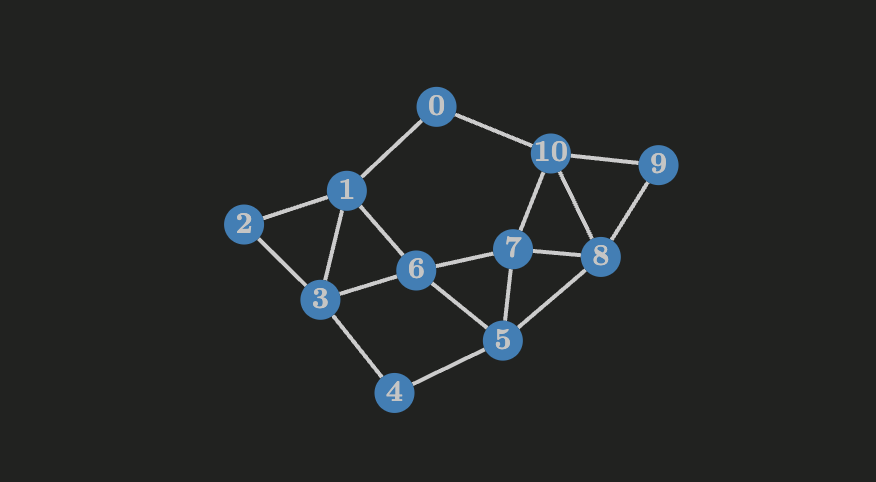
If a connected graph
Let
Minimum spanning tree
The purpose of finding a minimum spanning tree (MST) is to identify the subset of edges of a given connected, undirected graph that forms a tree, connecting all the vertices and having the minimum possible total edge weight.
Let
We are looking for a spanning tree of
- The MST is unique if the weights of all the edges are different.
- The maximum spanning tree is the tree whose weight is maximum among all spanning trees. It can be computed using the algorithm below by using the edges with the maximum weight instead of the ones with the minimum weight.
Kruskal’s algorithm
For a connected weighted graph
- For the first edge,
- For the second edge,
- For the third edge,
- We continue in this manner until a spanning tree is produced.
Let’s apply it to the weighted graph above. Sorting the edges in nondecreasing order, we have:
Some properties of the edges:
- The edge with the minimum cost is
- The edge with the minimum cost is now
- The next edge is
- The next edge is
- The next edge is
- There’s no need to do more iterations since the set is already a spanning tree.
struct edge {
int u, v, weight;
edge(int _u, int _v, int _w) {
u = _u; v = _v; weight = _w;
}
// custom sort
bool operator<(const edge &other) const {
return weight < other.weight;
}
};
vector<int> tree, size;
void initialize_sets(int n) {
tree.resize(n);
size.resize(n);
for (int i = 0; i < n; i += 1) {
tree[i] = i;
size[i] = 1;
}
}
int find_set(int element) {
if (element != tree[element]) {
tree[element] = find_set(tree[element]);
}
return tree[element];
}
void set_union(int x, int y) {
int rx, ry;
rx = find_set(x);
ry = find_set(y);
if (rx == ry) {
return;
}
if (rx > ry) {
size[rx] += size[ry];
tree[ry] = rx;
} else {
size[ry] += size[rx];
tree[rx] = ry;
}
}
/**
* An implementation of Kruskal's algorithm which computes
* the minimum spanning tree of a graph `G`
*
* Time complexity: O(m log m)
* Space complexity: O(m)
*
* @param {vector<vector<pair<int, int> > >} g The adjacency list representation
* of a graph `G`, each entry `g_{ij}` holds a pair which represents an edge
* (vertex, weight) which tells that there's an edge from `i` to `vertex`
* with weight `weight`
* @return {int} The weight of the MST
*/
int kruskal(vector<vector<pair<int, int> > > &g) {
int n = g.size();
vector<edge> edges;
for (int i = 0; i < n; i += 1) {
for (int j = 0; j < g[i].size(); j += 1) {
int v = g[i][j].first;
int weight = g[i][j].second;
edges.push_back(edge(i, v, weight));
}
}
initialize_sets(n);
sort(edges.begin(), edges.end());
int total = 0;
for (int i = 0; i < edges.size(); i += 1) {
int u = find_set(edges[i].u);
int v = find_set(edges[i].v);
if (u != v) {
set_union(u, v);
total += edges[i].weight;
}
}
return total;
}
Prim’s algorithm
For a connected weighted graph
- For an arbitrary vertex
- For subsequent edges
Prim in dense graphs
Let’s say we’re given the following problem:
Given
points in a plane, find the skeleton of minimum weight that connects them all.
This problem can be modeled as a graph of order
Implementation strategies:
- We need a data structure that keeps track of a single edge per vertex (space:
- After an arbitrary vertex
/**
* An implementation of Prim's algorithm which computes
* the minimum spanning tree of a dense graph `G`
*
* Time complexity: O(n^2)
* Space complexity: O(n)
*
* @param {vector<vector<int> >} g The adjacency matrix of `G`, each entry `a_{ij}`
* holds the weight of the edge connecting vertex `i` and vertex `j`, if this number
* is <= 0 then `i` and `j` are not adjacent
* @return {int} The weight of the MST
*/
int prim(vector<vector<int> > &g) {
int n = g.size();
int INF = 1e9;
int total = 0;
vector<bool> visited(n, false);
// holds the weight of the edge of minimum weight incident
// to the vertex `i`
vector<int> min_weight_edge(n, INF);
// (optional) holds the index of a vertex adjacent to the
// vertex `i` in the MST, note that the size of the MST is
// n - 1 so the first vertex won't store the mentioned index
vector<int> neighbor_selected(n, -1);
// pick the first node as the "arbitrary" node
min_weight_edge[0] = 0;
for (int i = 0; i < n; i += 1) {
int v = -1;
for (int j = 0; j < n; j += 1) {
// find the vertices which haven't been visited yet
// among them find a vertex with the minimum edge weight
if (!visited[j] && (v == -1 ||
min_weight_edge[j] < min_weight_edge[v])) {
v = j;
}
}
visited[v] = true;
total += min_weight_edge[v];
// update the minimum edge weight of all the vertices
// adjacent to `v`
for (int to = 0; to < n; to += 1) {
if (g[v][to] > 0 &&
g[v][to] < min_weight_edge[to]) {
min_weight_edge[to] = g[v][to];
// update the candidate neighbor of the vertex `to` to
// be `v` since it's connected with an edge
// of minimum weight among all the adjacent vertices to `to`
neighbor_selected[to] = v;
}
}
}
return total;
}
Prim in sparse graphs
Implementation strategies:
- We need a data structure that keeps track of a single edge per vertex (space:
- After an arbitrary vertex
- The red-black tree will hold at most
- There will be exactly
/**
* C++11
*
* An implementation of Prim's algorithm which computes
* the minimum spanning tree of a sparse graph `G` of order `n` and size `m`
*
* Time complexity: O(m log n)
* Space complexity: O(n)
*
* @param {vector<vector<pair<int, int> > >} g The adjacency list representation
* of a graph `G`, each entry `g_{ij}` holds a pair which represents an edge
* (vertex, weight) which tells that there's an edge from `i` to `vertex`
* with weight `weight`
* @return {int} The weight of the MST or a negative number if the graph
* wasn't connected
*/
int prim(vector<vector<pair<int, int> > > &g) {
int n = g.size();
int total = 0;
vector<bool> visited(n, false);
// holds the weight of the edge of minimum weight incident
// to the vertex `i`
vector<int> min_cost(n, INF);
// (optional) holds the index of a vertex adjacent to the
// vertex `i` in the MST, note that the size of the MST is
// n - 1 so the first vertex won't store the mentioned index
vector<int> neighbor(n, -1);
// the first node is the "arbitrary" node for the sake of the implementation
min_cost[0] = 0;
// (min weight, vertex)
set<pair<int, int> > q;
q.insert({0, 0});
while (!q.empty()) {
int v = q.begin()->second;
// the vertex `v` belongs to the MST and is adjacent
// to the vertex `neighbor[v]` with and edge
// of weight `weight`
total += q.begin()->first;
q.erase(q.begin());
visited[v] = true;
for (int i = 0; i < g[v].size(); i += 1) {
pair<int, int> &next = g[v][i];
// note that in the graph the first element is the neighbor vertex
// but in the set the first element is the edge weight
int to = next.first;
int weight = next.second;
if (!visited[to] && weight < min_cost[to]) {
q.erase({ min_cost[to], to });
min_cost[to] = weight;
neighbor[to] = v;
q.insert({ min_cost[to], to });
}
}
}
// check that every vertex has a min cost edge associated
for (int i = 0; i < n; i += 1) {
if (min_cost[i] == INF) {
return -1;
}
}
return total;
}
Number of spanning trees in a graph
Let
Then the number of spanning trees of
The matrix of cofactors is an
/**
* Given a square matrix M of size (n x n) this method
* computes the a matrix of size (n - 1) x (n - 1) by eliminating
* the elements belonging to the `row` row of M and the `col`
* column of M
*
* @param {vector<vector<int> > } m The square matrix
* @param {int} row The row to be ignored
* @param {int} col The column to be ignored
* @return {vector<vector<int> >} the value of the determinant
*/
vector<vector<int> > minor(vector<vector<int> > m, int row, int col) {
int n = m.size();
vector<vector<int> > t(n - 1, vector<int>(n - 1));
int trow = 0;
for (int i = 0; i < n; i += 1) {
if (i == row) {
continue;
}
int tcol = 0;
for (int j = 0; j < n; j += 1) {
if (j == col) {
continue;
}
t[trow][tcol] = m[i][j];
tcol += 1;
}
trow += 1;
}
return t;
}
/**
* Computes the determinant of an square matrix
*
* @param {vector<vector<int> > } m The square matrix
* @return {int} the value of the determinant
*/
int determinant(vector<vector<int> > m) {
int n = m.size();
if (n == 1) {
return m[0][0];
}
if (n == 2) {
return m[0][0] * m[1][1] - m[1][0] * m[0][1];
}
int result = 0;
for (int col = 0; col < n; col += 1) {
vector<vector<int> > t = minor(m, 0, col);
result += m[0][col] * pow(-1, col) * determinant(t);
}
return result;
}
/**
* Computes the number of spanning trees in an undirected graph `G`
*
* @param <vector<vector<int> > > g The adjacency matrix of `G`
* @return {int} The number of spanning trees
*/
int number_of_spanning_trees(vector<vector<int> > &g) {
int n = g.size();
vector<vector<int> > t = g;
for (int i = 0; i < n; i += 1) {
// -a_{ij} for elements that are not in the main diagonal
int degree = 0;
for (int j = 0; j < n; j += 1) {
if (i != j) {
t[i][j] *= -1;
if (t[i][j]) {
degree += 1;
}
}
}
// deg v_i for t[i][i]
t[i][i] = degree;
}
// compute the (0,0) cofactor
// c_{0, 0} = (-1)^{0 + 0} * determinant((0, 0) minor)
// = determinant((0, 0) minor)
return determinant(minor(t, 0, 0));
}
Number of spanning trees in a complete graph
Computing the number of spanning trees of a graph
The number of spanning trees of order
with a specific vertex set is .